
Image and Text Feature Based Multimodal Learning for Multi-Label
Classification of Radiology Images in Biomedical Literature
Md. Rakibul Hasan
a
, Md Rafsan Jani
b
and Md Mahmudur Rahman
c
Computer Science Department, Morgan State University, Baltimore, Maryland, U.S.A.
Keywords:
Biomedical Image Annotation, Image Retrieval, Multimodal Learning, ResNet50, ViT, CNN, DistilGPT2.
Abstract:
Biomedical images are crucial for diagnosing and planning treatments, as well as advancing scientific under-
standing of various ailments. To effectively highlight regions of interest (RoIs) and convey medical concepts,
annotation markers like arrows, letters, or symbols are employed. However, annotating these images with
appropriate medical labels poses a significant challenge. In this study, we propose a framework that leverages
multimodal input features, including text/label features and visual features, to facilitate accurate annotation
of biomedical images with multiple labels. Our approach integrates state-of-the-art models such as ResNet50
and Vision Transformers (ViT) to extract informative features from the images. Additionally, we employ Gen-
erative Pre-trained Distilled-GPT2 (Transformer based Natural Language Processing architecture) to extract
textual features, leveraging their natural language understanding capabilities. This combination of image and
text modalities allows for a more comprehensive representation of the biomedical data, leading to improved
annotation accuracy. By combining the features extracted from both image and text modalities, we trained a
simplified Convolutional Neural Network (CNN) based multi-classifier to learn the image-text relations and
predict multi-labels for multi-modal radiology images. We used ImageCLEFmedical 2022 and 2023 datasets
to demonstrate the effectiveness of our framework. This dataset likely contains a diverse range of biomedical
images, enabling the evaluation of the framework’s performance under realistic conditions. We have achieved
promising results with the F1 score of 0.508. Our proposed framework exhibits potential performance in an-
notating biomedical images with multiple labels, contributing to improved image understanding and analysis
in the medical image processing domain.
1 INTRODUCTION
The advent of digital technology in the biomedical
field has led to an exponential increase in the vol-
ume of available radiology images and associated tex-
tual data within biomedical literature. This wealth
of information, while invaluable, presents significant
challenges in terms of efficient classification and re-
trieval (Dhawan, 2011). As a result, developing tool
for annotating and classifying medical images to as-
sist users, such as patients, researchers, general prac-
titioners, and clinicians, in finding pertinent and help-
ful information is being considered as the active re-
search domain in the biomedical sector (Demner-
Fushman et al., 2009). The paper titled ”Image and
Text Feature-based Multimodal Learning for Multi-
label Classification of Radiology Images in Biomed-
a
https://orcid.org/0000-0002-6179-2238
b
https://orcid.org/0000-0001-7304-087X
c
https://orcid.org/0000-0003-0405-9088
ical Literature” addresses the challenge of contribut-
ing in this domain by exploring the integration of both
image and text features in the multi-classification pro-
cess.
Radiology images, such as X-rays, Computed
Tomography (CT) scans, and Magnetic Resonance
Imaging (MRI), are a cornerstone of medical diag-
nostics and research, offering vital insights into var-
ious medical conditions (Azam et al., 2022). How-
ever, the sheer volume and complexity of these im-
ages, coupled with the accompanying textual descrip-
tions, necessitate advanced methods for effective or-
ganization and retrieval (Rahman et al., 2015). Tra-
ditional approaches often rely heavily on either text-
based or image-based features, neglecting the poten-
tial synergy of a multimodal approach (Ritter et al.,
2011). However, this paper explores the approach of
multimodal learning by exploiting different state-of-
the art deep learning frameworks to leverage both im-
age and text features for the multi-label classification
Hasan, M., Jani, M. and Rahman, M.
Image and Text Feature Based Multimodal Learning for Multi-Label Classification of Radiology Images in Biomedical Literature.
DOI: 10.5220/0012438400003657
Paper published under CC license (CC BY-NC-ND 4.0)
In Proceedings of the 17th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2024) - Volume 2, pages 679-686
ISBN: 978-989-758-688-0; ISSN: 2184-4305
Proceedings Copyright © 2024 by SCITEPRESS – Science and Technology Publications, Lda.
679

of radiology images in biomedical literature. By in-
tegrating visual cues from the images with contextual
information derived from textual data, our approach
aims to enhance the accuracy and efficiency of classi-
fication tasks. This is particularly crucial in the con-
text of biomedical literature, where the precise cate-
gorization of images is essential for aiding research,
clinical decision-making, and educational purposes.
To achieve the aim of this research paper, we uti-
lized the medical image dataset obtained from the
ImageCLEFmedical Caption Tasks of 2022 (Ionescu
et al., 2022) and 2023 (Ionescu et al., 2023), (Barr
´
on-
Cedeno et al., 2023). The dataset of the year 2022
comprises 83,275 training images and 7,645 valida-
tion images. The 2023 dataset also includes 60,918
training images, and 10,437 validation images. The
medical images on these datasets are multi-modal
which includes X-ray, CT scan, MRI, Ultrasound,
PET scan, Angiogram, and other types of radiology
images. Each of the images pertaining to the train
and validation sets are associated with captions and
concepts. The medical concepts were presented fol-
lowing the UMLS format. Later on, 2,125 Concept
Unique Identifiers (CUIs) were employed to repre-
sent the UMLS terminologies which results each im-
age having multiple CIUs or labels. The test dataset is
not used or reported here because it only has the im-
ages, corresponding concepts and captions were kept
hidden for the competition purposes. It’s important to
note that both of these datasets are the updated and
extended version of the ROCO (Radiology Objects in
Context) image dataset collected from various open
access journals in PubMed (Pelka et al., 2018).
For our research, we have used 2023 CLEF train-
ing and validation images as our training and testing
purposes, correspondingly. As a result, 60,918 im-
ages are used for train our intended model and 10,437
images are used to test our trained model. On the
other hand, 16,358 images selected from the training
and validation image sets of the 2022 year are used
for validation purpose in this research. The images of
the 2022 dataset having CUIs not used in 2023 are ex-
cluded from our validation dataset. As a result, the to-
tal number of unique labels is kept within the number
of 2,125. Moreover, the possibility of repetitive use of
same images in training, validation, and test datasets
are minimized. In total 86,993 images are used in our
research, whereas the ratio of training, validation, and
test image-label pairs are approximately 70%, 20%,
and 10%, respectively.
Top three of the most frequent CUIs found on
the train and validation dataset are ’C0040405’ (Fre-
quency: 24,695), ’C1306645’ (Frequency: 19,833),
and ’C0024485’ (Frequency: 11,554); correspond-
ing UMLS are ’Magnetic Reasonance Imaging’,
’Anterior-Posterior’, and ’Angiogram’. Each image
has average five multi-labels or CUIs and maximum
number of labels for an image found in the dataset
is fifty. In addition, the set of the image captions
has 23,237 corpus of words. The maximum number
of words in a caption for an image is found as 316,
however the 99% images are having 90 or less than
number of words as caption. Along with the respec-
tive multi-labels represented as CUIs/UMLS and cap-
tions, we have prepared processed caption for each
image based on the UMLS. For example, an image
has the above mentioned three UMLS, then the pro-
cessed caption for that image is prepared by placing
”This image shows” at the beginning and followed the
UMLS sequentially. This is worthy to mention that in
the original dataset the UMLS are the keywords de-
rived from the captions, as a result the processed cap-
tion works better instead of pre-processing the origi-
nal captions with some standard natural language pro-
cessing techniques like removing stop words, special
characters, numeric values, and converting to lower
case, etc.
Figure 1 shows an instance from the dataset we
have used in our study.
Figure 1: A sample MRI image from our test dataset
(CC BY-NC [Murvelashvili et al. (2021)]). The corre-
sponding CUIs: [‘C0024485’, ‘C0449900’, ‘C0006104’,
‘C0014008’], UMLS: [‘Magnetic Resonance Imaging’,
‘Contrast used’, ‘Brain’, ‘Empty Sella Syndrome’], Pro-
cessed Caption: ‘this image shows magnetic resonance
imaging contrast used brain empty sella syndrome’, and
Original Caption: ‘Contrast-enhanced T1-weighted sagittal
image of the brain 1 month after initial presentation. The
arrow shows a mostly empty sella.’.
2 RELATED WORKS
Multi-label classification of medical images is a task
that involves assigning multiple labels or categories to
an image, allowing for a more comprehensive and nu-
anced representation of the content within the image.
This task is particularly relevant in the biomedical
HEALTHINF 2024 - 17th International Conference on Health Informatics
680

field, where images often contain multiple character-
istics, findings, or abnormalities that need to be accu-
rately identified and labeled. The objective of multi-
label classification is to develop a model that can fore-
tell the pertinent labels for a given input, which could
be radiology reports, images, or any other kind of data
(Zhang and Zhou, 2013). Each instance in the labeled
dataset used to train the model has a set of labels at-
tached to it.
The recent progress relavant to the task of multi-
label classification mainly evolves around two types
of deep learning models based on Convolutional Neu-
ral Network (CNN) approach and Transformer ap-
proach. Several state-of-the-art (SOTA) CNN archi-
tectures have been employed for multi-label classifi-
cation of medical images. To tackle the challenges as-
sociated with multi-label classification, various prac-
tices and methodologies have been adopted. One
approach involves utilizing pre-trained CNN mod-
els, trained on large-scale generic image datasets, and
fine-tuning them on specific medical image datasets
(Tajbakhsh et al., 2016). These includes the tech-
niques of applying ensemble method, transfer learn-
ing, and using pre-trained models/weights on com-
paratively larger image sets. Transfer learning, as
the most prominent one, involves transferring knowl-
edge learned from one domain to another, has been
effective in improving the performance of models
when training data is limited.Standard CNN archi-
tectures such as ResNet (He et al., 2016), Inception-
Net (Szegedy et al., 2016), EfficientNet (Tan and Le,
2019), VGGNet (Simonyan and Zisserman, 2014),
and DenseNet (Huang et al., 2017) are widely used for
this task due to their ability to capture intricate visual
features and patterns from images. Moreover, each of
these architectures comes with some unique features
to demonstrate their corresponding ability of image
analysis. In (Hasan et al., 2023), (Yeshwanth et al.,
2023) workshop notes, the participants of the Image-
CLEF2023 medical tasks, the use of DenseNet121
were demonstrated for the task of concept detection or
multi-label classification using the CLEF23 dataset.
Futhermore, in (Kaliosis et al., 2023) and (Shinoda
et al., 2023), EfficientNetV2 and the fusion of Effi-
cientNet with DenseNet were utilized for multi-label
classification. VGG16 and a customized CNN archi-
tecture nmaed as ConceptNet were employed by (Rio-
Torto et al., 2023) and (Mohamed and Srinivasan,
2023), respectively.
Overall, the core idea of Convolution is excellent
for tasks where local spatial or temporal relationships
are key, and its efficiency and simplicity make it a
staple in many applications. Attention, on the other
hand, shines in scenarios where the model needs to
dynamically focus on different parts of the input, cap-
turing long-range interactions and dependencies ef-
fectively (He et al., 2016), (Xu et al., 2015). More
precisely, in the context of multi-modal medical im-
age analysis to extract image features and establish
long range dependencies between the modalities at-
tention based Transformer architectures are thriving
in the recent times (Dai et al., 2021). Vision Trans-
former (ViT) (Dosovitskiy et al., 2020), Swin Trans-
former (Liu et al., 2021), and Data-efficient Image
Transformer (DeIT) (Touvron et al., 2021) are be-
ing widely used in medical image classification tasks
(Manzari et al., 2023), (Okolo et al., 2022).
3 MODEL IMPLEMENTATION
The visual representations that are extracted from im-
ages, generally utilizing methods like Convolution
and/or Transformer based architectures, are referred
to as image features. Effective image analysis and
classification are made possible by these features,
which capture significant visual patterns and char-
acteristics contained in the images (Liu and Deng,
2015). Contrarily, text features entail the display
of text-based content such image captions, radiolog-
ical reports, or clinical notes. Deep leaning archi-
tectures specialized on processing natural or scien-
tific languages/texts can be used to extract text fea-
tures, which convert the text into numerical represen-
tations which also convey important insights about
the corresponding medical images besides image fea-
tures (Pennington et al., 2014). As a result, the ap-
proach of incorporating textual information with vi-
sual features, such as radiology reports or image cap-
tions, alongside the images for multi-modal learning
is adopted in this research (Yao et al., 2019). By com-
bining both visual and textual features, models can
leverage the complementary information from both
modalities to improve classification accuracy. Multi-
modal learning mixes text and visual features to take
advantage of complimentary data from several modal-
ities. Multimodal models seek to enhance the per-
formance of tasks like picture classification, object
recognition, or image captioning by combining the vi-
sual and linguistic characteristics. This strategy uses
both visual and semantic cues to provide a more thor-
ough grasp of the material (Frome et al., 2013).
The approach of connecting image and text fea-
tures is recently being applied in the field of auto-
mated caption generation for images. Such as CLIP
(Contrastive Language–Image Pre-training) (Radford
et al., 2021) model compresses both image and
text features to establish image-to-text connection in
Image and Text Feature Based Multimodal Learning for Multi-Label Classification of Radiology Images in Biomedical Literature
681
zero-shot manner. Another state-of-the-art model
BLIP (Bootstrapping Language-Image Pre-training)
(Li et al., 2022) works at the same of unifying both
vision and language features to generate captions.
In our implemented model due to the constraint
of computational resources, we have tried to exploit
the prediction prowess of pre-trained SOTA CNN and
Transformer based models followed by a simple CNN
classifier. The pre-trained models we have used here
are ResNet50, ViT32, and DistilGPT2.
ResNet (Residual Neural Network) (He et al.,
2016) is a groundbreaking neural network architec-
ture that has addressed the vanishing gradient prob-
lem of training very deep neural networks by intro-
ducing residual connections, enabling the training of
models with hundreds or even thousands of layers.
The residual block is the key building block of the
ResNet. Each residual block consists of a sequence
of convolutional layers, batch normalization, and rec-
tified linear unit (ReLU) activations. The innova-
tion lies in the introduction of skip connections, also
known as shortcut connections, which allow the net-
work to learn residual mappings (Yu et al., 2018). The
skip connections in ResNet enable the direct propa-
gation of information from one layer to subsequent
layers.
Vision Transformer (ViT) extends the Trans-
former model, which was initially developed for nat-
ural language processing, to the field of computer vi-
sion. ViT analyzes images as a series of flattened
patches rather than the traditional grid-based convo-
lutional neural networks (CNNs) that do the same.
Each of the fixed-size patches (Dosovitskiy et al.,
2020) that make up ViT’s input image is linearly pro-
jected to a lower-dimensional representation. Posi-
tional embeddings are provided to incorporate spa-
tial information, enabling the model to understand
the relative placements of patches. The patch em-
beddings are then passed via a Transformer encoder,
which consists of several feed-forward neural net-
works and self-attention layers. A crucial element
of the Transformer encoder (Touvron et al., 2021) is
self-attention, which enables the model to recognize
inter-dependencies and focus on various areas of the
image while processing. ViT learns complex local
and global context representations by simultaneously
monitoring all patches. The computation of attention
weights between pairs of patches is carried out by the
self-attention mechanism. Long-range dependencies
are now simpler to represent, and patches can now
communicate with one another. In order to conduct
image classification tasks with an accurate prediction
of the class labels, a classification head is added af-
ter the patch embeddings have been processed by the
Transformer encoder. ViT is frequently trained using
supervised learning on big datasets during the train-
ing phase, and the model parameters are optimized
via backpropagation and gradient-based optimization.
A state-of-the-art language model that excels at
natural language processing (NLP) tasks is called
GPT (Generative Pre-trained Transformer). Trans-
former served as the foundation for GPT, which em-
ploys self-attention processes to identify connections
and complicated language patterns in textual data
(Radford et al., 2019). An important variation of
GPT2 is Distilled-GPT2 or DistilGPT2 developed by
Hugging Face (Sanh et al., 2019). DistilGPT-2 can be
used in similar applications as GPT-2, including text
generation, chatbots, content creation, and more. Its
smaller size makes it particularly useful in scenarios
where deploying a full GPT-2 model would be im-
practical due to resource constraints. Moreover, Dis-
tilGPT2 is believed to be more efficient in generat-
ing scientific texts. The self-attention (Vaswani et al.,
2017) method, which enables the model to concen-
trate on key portions of the input sequence while con-
structing each word, helps with this.
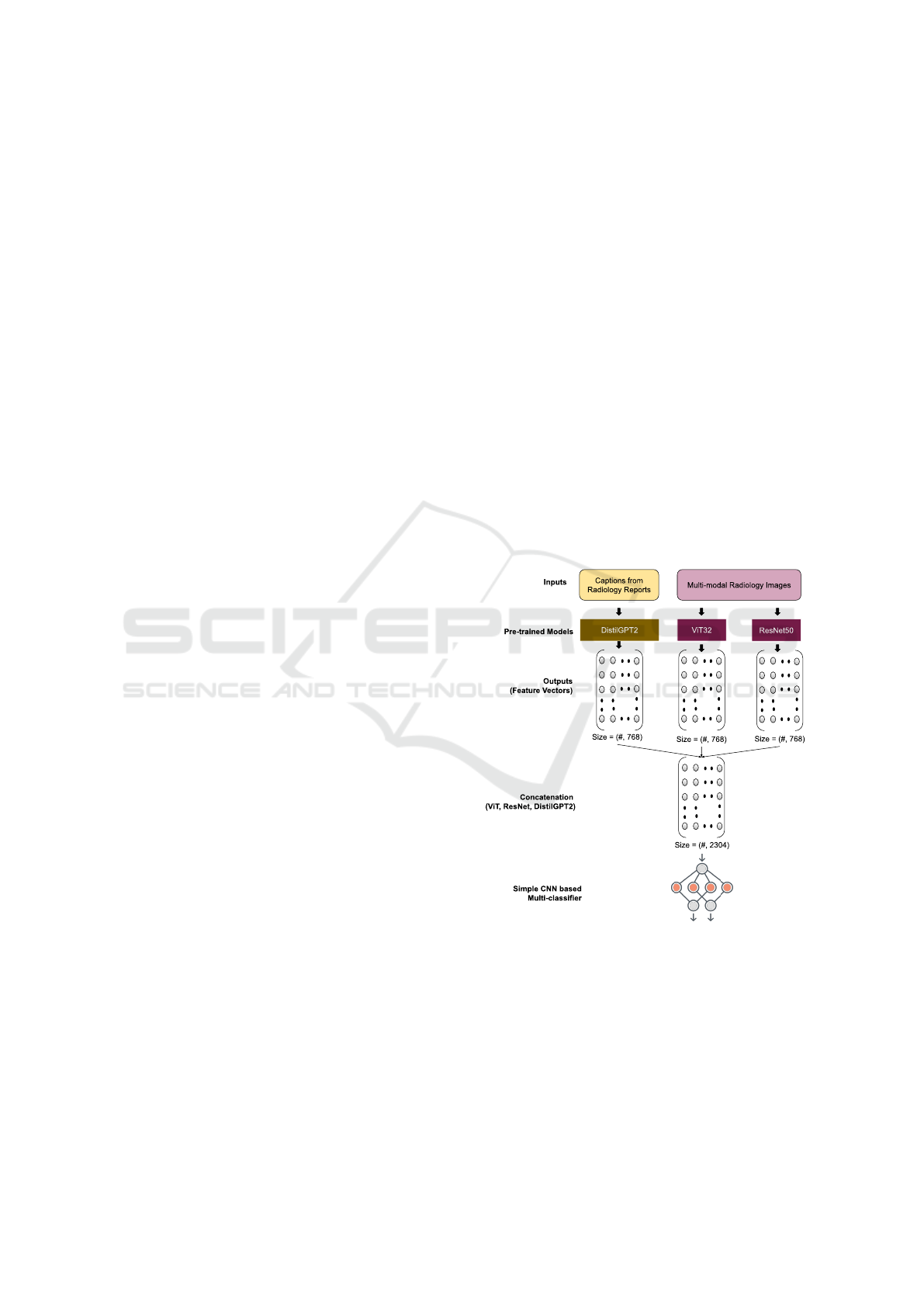
Figure 2: The Implmented Model.
According to Figure 2, we have utilized the pre-
trained ResNet50 and ViT32 architectures to extract
the visual features of the images. Each image will
have the feature vector of size=(768,). On the other
hand, the text feature vectors are extracted based on
the processed captions of each image by using the
DistilGPT2 pre-trained model, which has the embed-
ding size=(768,) for each caption. After getting the
visual and textual features a Concatenation() tech-
HEALTHINF 2024 - 17th International Conference on Health Informatics
682
nique is applied to merge the vectors which gener-
ates size=(2304,). Later on, a simplified CNN based
multi-classifier is employed to feed the concatenated
feature vectors into it and predict multi-labels or con-
cepts for the images from the test dataset using 2,125
unique classes.
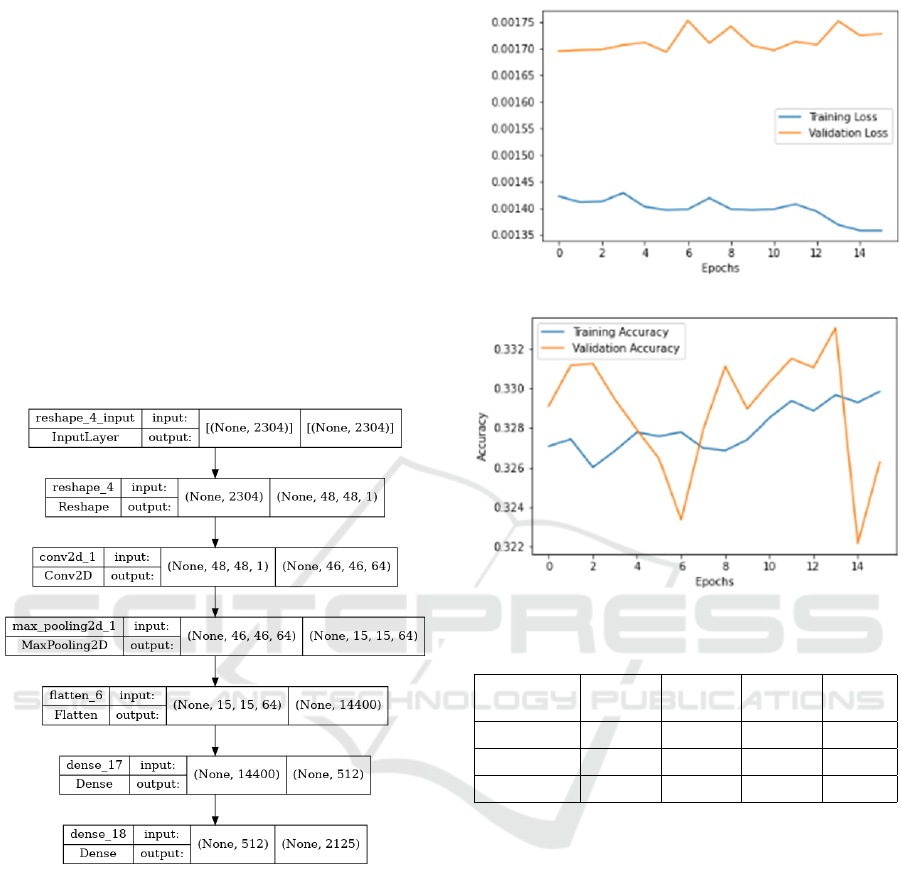
Figure 3 depicts the custom architecture of
our simplified CNN multi-classifier, which consists
of Reshape layer at the beginning to convert the
size=(2304,) to size=(48x48x1) required for the next
Convolutional2D layer, then the next sequence of lay-
ers is MaxPooling2D ⇒ Flatten ⇒ Dense layer. Fi-
nally, a Dense layer with ‘sigmoid’ activation func-
tion is implemented to predict the multi-labels from
the set of 2,215 labels. Furthermore, the Binary
Crossentropy loss function is used to predict the prob-
ability of each labels for a test image.
Figure 3: The Simplified CNN based Multi-classification.
4 EXPERIMENTS & RESULTS
Figure 4 depicts the loss and accuracy both for the
training and validation phases. Here, loss represents
the discrepancy between the predicted labels and the
true labels. On the other hand, accuracy is a met-
ric that measures the proportion of correctly classified
samples out of the total number of samples.
Table 1 shows the key performance metrics of
model in predicting multi-labels for the test dataset.
Precision is the measure of the model’s ability to
correctly identify positive samples out of all the sam-
ples predicted as positive. Recall is the measure of
Figure 4: Loss and Accuracy.
Table 1: Key Performance Metrics.
Accuracy Precision Recall F1-
score
Training 0.329 - - -
Validation 0.326 - - -
Testing - 0.697 0.447 0.508
the model’s ability to correctly identify positive sam-
ples out of all the true positive samples. The F1 score
combines precision and recall providing a balanced
evaluation of the model’s performance, considering
both false positives and false negatives. We have also
conducted an investigation of predicting multi-labels
without using text features, which results less perfor-
mance than our proposed model of using both visual
and textual features. More precisely, the F1 score is
found 0.31 (approximately) for the model of exclud-
ing text features. On the other hand, comparing our
results with the CLEF2023 challenge participant’s re-
sults will be misleading, because the F1 scores of
those participants were calculated by the organizer us-
ing the test dataset where the associated multi-labels
were not disclosed.
However, Figure 5-7 depicts the predicted multi-
labels for some of the test images used in our research
in comparison to the actual ground truth labels.
Image and Text Feature Based Multimodal Learning for Multi-Label Classification of Radiology Images in Biomedical Literature
683
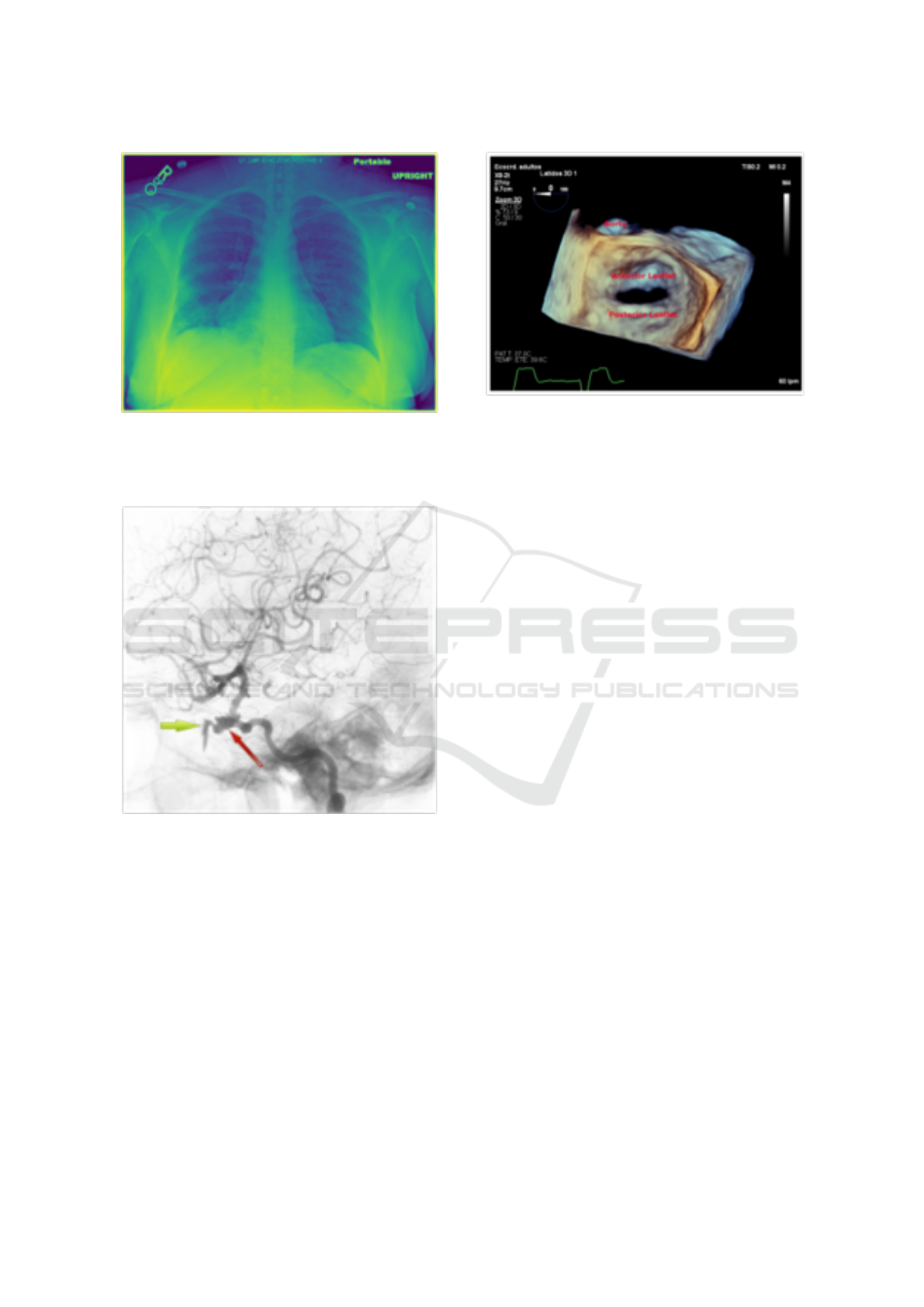
Figure 5: Random Test Image (CC BY-NC [Ogamba et al.
(2021)]); Ground Truth CUIs: [‘C1306645’, ‘C0817096’,
‘C1999039’, ‘C0039985’]; Predicted CUIs: [‘C1306645’,
‘C1999039’, ‘C1306645’, ‘C0032285’].
Figure 6: Random Test Image (CC BY [Muacevic
et al. (2021)]); Ground Truth CUIs: [‘C0002978’,
‘C0002940’, ‘C0226156’, ‘C0582802’, ‘C0007276’]; Pre-
dicted CUIs: [’C0040405’, ’C0817096’,‘C0002940’,
‘C0226156’, ‘C0582802’].
5 CONCLUSION
Valuable information is provided by harnessing the
often overlooked textual and visual content, going
beyond traditional databases. Future efforts include
generating more training data and building advanced
information retrieval systems with a fusion model.
However, the models used in the study face limi-
tations in predicting with higher accuracy followed
by an well-defined pre-processing techniques of im-
Figure 7: Random Test Image (CC BY [Ruiz et al.
(2021)]); Ground Truth CUIs: [‘C0026264’, ‘C0225860’,
‘C0003483’]; Predicted CUIs: [‘C0026264’, ‘C0002978’,
‘C0456598’, ‘C0190010’,‘C0003483’].
ages using Keras networks for multi-label classifica-
tion. Future work aims to overcome this by focusing
on deep learning-based object detection. The impact
of this research is substantial for applications such
as digital libraries and image search engines, which
demand efficient techniques for image categorization
and access.
ACKNOWLEDGEMENTS
This work is supported by the National Science Foun-
dation (NSF) grant (ID: 2131307) under CISE-MSI
program.
REFERENCES
Azam, M. A., Khan, K. B., Salahuddin, S., Rehman, E.,
Khan, S. A., Khan, M. A., Kadry, S., and Gandomi,
A. H. (2022). A review on multimodal medical im-
age fusion: Compendious analysis of medical modal-
ities, multimodal databases, fusion techniques and
quality metrics. Computers in biology and medicine,
144:105253.
Barr
´
on-Cedeno, A., Da San Martino, G., Esposti, M. D.,
Faggioli, G., Ferro, N., Hanbury, A., Macdonald, C.,
Pasi, G., Potthast, M., and Sebastiani, F. (2023). Re-
port on the 13th conference and labs of the evaluation
forum (clef 2022) experimental ir meets multilingual-
ity, multimodality, and interaction. In ACM SIGIR Fo-
rum, volume 56, pages 1–15. ACM New York, NY,
USA.
Dai, Y., Gao, Y., and Liu, F. (2021). Transmed: Transform-
ers advance multi-modal medical image classification.
Diagnostics, 11(8):1384.
Demner-Fushman, D., Antani, S., Simpson, M., and
HEALTHINF 2024 - 17th International Conference on Health Informatics
684

Thoma, G. R. (2009). Annotation and retrieval of clin-
ically relevant images. international journal of medi-
cal informatics, 78(12):e59–e67.
Dhawan, A. P. (2011). Medical image analysis. John Wiley
& Sons.
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn,
D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer,
M., Heigold, G., Gelly, S., et al. (2020). An image is
worth 16x16 words: Transformers for image recogni-
tion at scale. arXiv preprint arXiv:2010.11929.
Frome, A., Corrado, G. S., Shlens, J., Bengio, S., Dean,
J., Ranzato, M., and Mikolov, T. (2013). Devise: A
deep visual-semantic embedding model. Advances in
neural information processing systems, 26.
Hasan, M. R., Layode, O., and Rahman, M. (2023).
Concept detection and caption prediction in image-
clefmedical caption 2023 with convolutional neural
networks, vision and text-to-text transfer transform-
ers. In CLEF2023 Working Notes, CEUR Workshop
Proceedings, Thessaloniki, Greece. CEURWS.org.
He, K., Zhang, X., Ren, S., and Sun, J. (2016). Deep resid-
ual learning for image recognition. In Proceedings of
the IEEE conference on computer vision and pattern
recognition, pages 770–778.
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger,
K. Q. (2017). Densely connected convolutional net-
works. In Proceedings of the IEEE conference on
computer vision and pattern recognition, pages 4700–
4708.
Ionescu, B., M
¨
uller, H., Dr
˘
agulinescu, A. M., Popescu, A.,
Idrissi-Yaghir, A., Garc
´
ıa Seco de Herrera, A., An-
drei, A., Stan, A., Stor
˚
as, A. M., Abacha, A. B., et al.
(2023). Imageclef 2023 highlight: Multimedia re-
trieval in medical, social media and content recom-
mendation applications. In European Conference on
Information Retrieval, pages 557–567. Springer.
Ionescu, B., M
¨
uller, H., P
´
eteri, R., R
¨
uckert, J., Abacha,
A. B., de Herrera, A. G. S., Friedrich, C. M., Bloch,
L., Br
¨
ungel, R., Idrissi-Yaghir, A., et al. (2022).
Overview of the imageclef 2022: Multimedia retrieval
in medical, social media and nature applications.
In International Conference of the Cross-Language
Evaluation Forum for European Languages, pages
541–564. Springer.
Kaliosis, P., Moschovis, G., Charalambakos, F., Pavlopou-
los, J., and Androutsopoulos, I. (2023). Aueb
nlp group at imageclefmedical caption 2023. In
CLEF2023 Working Notes, CEUR Workshop Pro-
ceedings, Thessaloniki, Greece. CEUR-WS.org.
Li, J., Li, D., Xiong, C., and Hoi, S. (2022). Blip:
Bootstrapping language-image pre-training for unified
vision-language understanding and generation. In In-
ternational Conference on Machine Learning, pages
12888–12900. PMLR.
Liu, S. and Deng, W. (2015). Very deep convolutional
neural network based image classification using small
training sample size. In 2015 3rd IAPR Asian confer-
ence on pattern recognition (ACPR), pages 730–734.
IEEE.
Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z., Lin,
S., and Guo, B. (2021). Swin transformer: Hierar-
chical vision transformer using shifted windows. In
Proceedings of the IEEE/CVF international confer-
ence on computer vision, pages 10012–10022.
Manzari, O. N., Ahmadabadi, H., Kashiani, H., Shokouhi,
S. B., and Ayatollahi, A. (2023). Medvit: a ro-
bust vision transformer for generalized medical image
classification. Computers in Biology and Medicine,
157:106791.
Mohamed, S. S. N. and Srinivasan, K. (2023). Ssn
mlrg at caption 2023: Automatic concept detection
and caption prediction using conceptnet and vision
transformer. In CLEF2023 Working Notes, CEUR
Workshop Proceedings, Thessaloniki, Greece. CEUR-
WS.org.
Okolo, G. I., Katsigiannis, S., and Ramzan, N. (2022). Ievit:
An enhanced vision transformer architecture for chest
x-ray image classification. Computer Methods and
Programs in Biomedicine, 226:107141.
Pelka, O., Koitka, S., R
¨
uckert, J., Nensa, F., and Friedrich,
C. M. (2018). Radiology objects in context (roco):
a multimodal image dataset. In Intravascular Imag-
ing and Computer Assisted Stenting and Large-Scale
Annotation of Biomedical Data and Expert Label
Synthesis: 7th Joint International Workshop, CVII-
STENT 2018 and Third International Workshop, LA-
BELS 2018, Held in Conjunction with MICCAI 2018,
Granada, Spain, September 16, 2018, Proceedings 3,
pages 180–189. Springer.
Pennington, J., Socher, R., and Manning, C. D. (2014).
Glove: Global vectors for word representation. In
Proceedings of the 2014 conference on empirical
methods in natural language processing (EMNLP),
pages 1532–1543.
Radford, A., Kim, J. W., Hallacy, C., Ramesh, A., Goh, G.,
Agarwal, S., Sastry, G., Askell, A., Mishkin, P., Clark,
J., et al. (2021). Learning transferable visual models
from natural language supervision. In International
conference on machine learning, pages 8748–8763.
PMLR.
Radford, A., Wu, J., Child, R., Luan, D., Amodei, D.,
Sutskever, I., et al. (2019). Language models are un-
supervised multitask learners. OpenAI blog, 1(8):9.
Rahman, M. M., Antani, S. K., Demner-Fushman, D., and
Thoma, G. R. (2015). Biomedical image representa-
tion approach using visualness and spatial information
in a concept feature space for interactive region-of-
interest-based retrieval. Journal of Medical Imaging,
2(4):046502–046502.
Rio-Torto, I., Patr
´
ıcio, C., Montenegro, H., Gonc¸alves, T.,
and Cardoso, J. S. (2023). Detecting concepts and
generating captions from medical images: Contribu-
tions of the vcmi team to imageclefmedical caption
2023. In CLEF2023 Working Notes, Thessaloniki,
Greece. CEUR-WS.org, CEUR Workshop Proceed-
ings.
Ritter, F., Boskamp, T., Homeyer, A., Laue, H., Schwier,
M., Link, F., and Peitgen, H.-O. (2011). Medical im-
age analysis. IEEE pulse, 2(6):60–70.
Sanh, V., Debut, L., Chaumond, J., and Wolf, T. (2019).
Image and Text Feature Based Multimodal Learning for Multi-Label Classification of Radiology Images in Biomedical Literature
685

Distilbert, a distilled version of bert: smaller, faster,
cheaper and lighter. In NeurIPS EMC
2
Workshop.
Shinoda, H., Aono, M., Asakawa, T., Shimizu, K., Ko-
moda, T., and Togawa, T. (2023). Kde lab at im-
ageclefmedical caption 2023. In CLEF2023 Working
Notes, CEUR Workshop Proceedings, Thessaloniki,
Greece. CEUR-WS.org.
Simonyan, K. and Zisserman, A. (2014). Very deep con-
volutional networks for large-scale image recognition.
arXiv preprint arXiv:1409.1556.
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wo-
jna, Z. (2016). Rethinking the inception architecture
for computer vision. In Proceedings of the IEEE con-
ference on computer vision and pattern recognition,
pages 2818–2826.
Tajbakhsh, N., Shin, J. Y., Gurudu, S. R., Hurst, R. T.,
Kendall, C. B., Gotway, M. B., and Liang, J. (2016).
Convolutional neural networks for medical image
analysis: Full training or fine tuning? IEEE trans-
actions on medical imaging, 35(5):1299–1312.
Tan, M. and Le, Q. (2019). Efficientnet: Rethinking model
scaling for convolutional neural networks. In Interna-
tional conference on machine learning, pages 6105–
6114. PMLR.
Touvron, H., Cord, M., Sablayrolles, A., Synnaeve, G., and
J
´
egou, H. (2021). Going deeper with image transform-
ers. In Proceedings of the IEEE/CVF international
conference on computer vision, pages 32–42.
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones,
L., Gomez, A. N., Kaiser, Ł., and Polosukhin, I.
(2017). Attention is all you need. Advances in neural
information processing systems, 30.
Xu, K., Ba, J., Kiros, R., Cho, K., Courville, A., Salakhudi-
nov, R., Zemel, R., and Bengio, Y. (2015). Show, at-
tend and tell: Neural image caption generation with
visual attention. In International conference on ma-
chine learning, pages 2048–2057. PMLR.
Yao, J., Zhu, X., and Huang, J. (2019). Deep multi-
instance learning for survival prediction from whole
slide images. In Medical Image Computing and Com-
puter Assisted Intervention–MICCAI 2019: 22nd In-
ternational Conference, Shenzhen, China, October
13–17, 2019, Proceedings, Part I 22, pages 496–504.
Springer.
Yeshwanth, V., P, P., and Kalinathan, L. (2023). Concept de-
tection and image caption generation in medical imag-
ing. In CLEF2023 Working Notes, CEUR Workshop
Proceedings, Thessaloniki, Greece. CEUR-WS.org.
Yu, X., Yu, Z., and Ramalingam, S. (2018). Learning strict
identity mappings in deep residual networks. In Pro-
ceedings of the IEEE Conference on Computer Vision
and Pattern Recognition, pages 4432–4440.
Zhang, M.-L. and Zhou, Z.-H. (2013). A review on
multi-label learning algorithms. IEEE transactions on
knowledge and data engineering, 26(8):1819–1837.
HEALTHINF 2024 - 17th International Conference on Health Informatics
686
